Fusion Antibodies
End-to-end antibody development partner
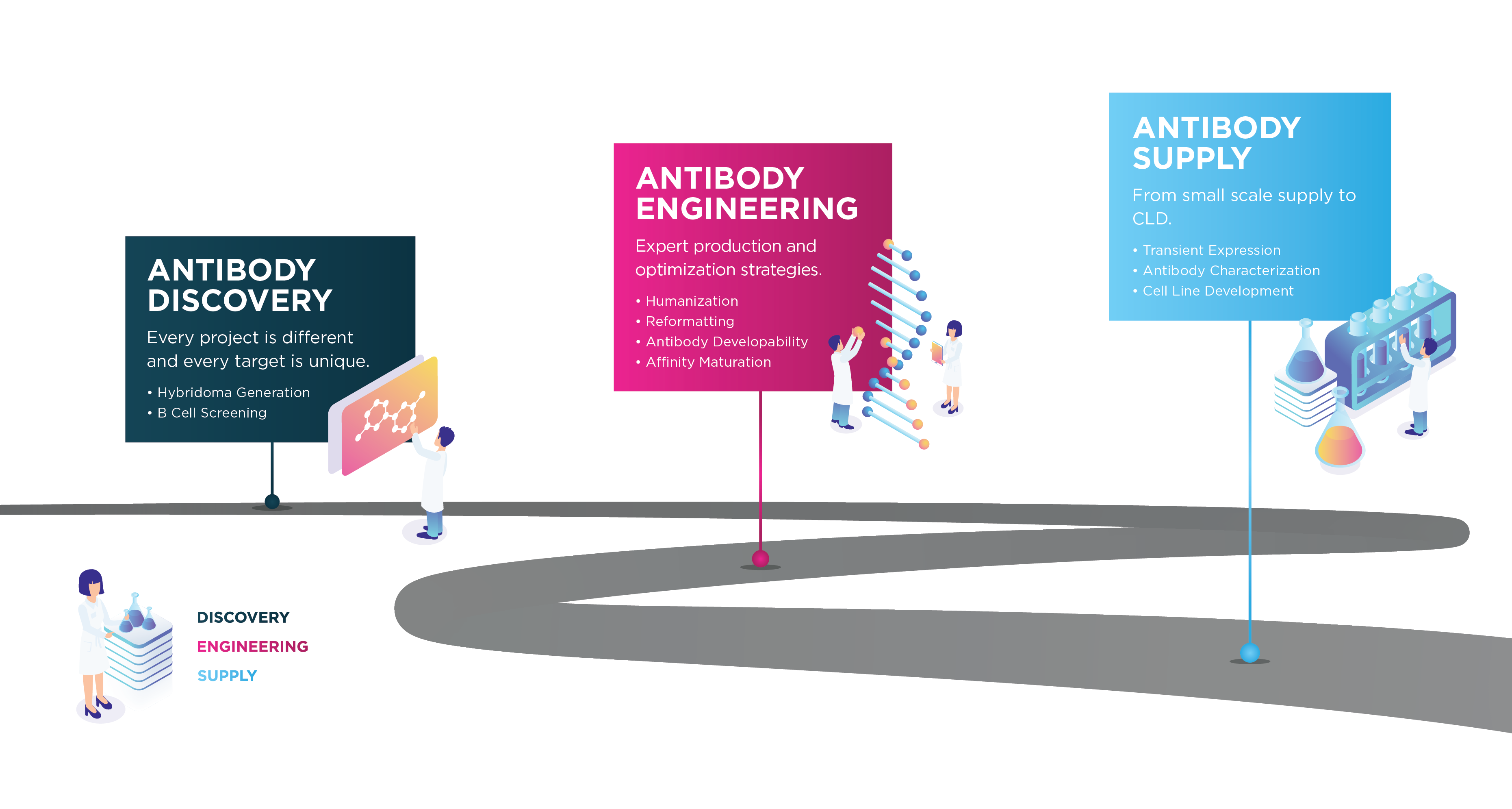
Progressing your antibody to the clinic is challenging, but we understand the hurdles. We offer a complete end-to-end service from antigen design to cell line development.
We work in partnership as an extension of your team to deliver the project requirements while also considering long term goals of manufacturability and commercialization.