The challenges of phage display for affinity maturation
Phage display has had a good run. After bursting onto the antibody engineering scene in the 1990s, phage display rapidly disrupted the field. The in vitro high throughput screening and selection offered by phage display hit the jackpot in 2002 with the approval of adalimumab (Humira®), which we now know as the world’s best-selling therapeutic antibody.
However phage display’s crown has been slowly slipping. Despite early promise, only 10 of the 100 or so FDA-licensed monoclonal antibodies were developed using phage display (1, 2). So what are the pitfalls of phage?
Antibodies on display in phage
Phage, or bacteriophage to give them their full name, selectively infect bacteria and hitchhike the bacterial machinery to replicate themselves. Biologists in turn have hijacked phage by inserting genetic code into the phage which is then manufactured into a protein by the bacterial hosts’ protein expression system and displayed on the phage outside coat. This link between the genotype (the genetic sequence inserted into the phage) and the phenotype (the antibody manufactured based on the code) is the most important principle of phage display.
Antibody engineers exploit this system for antibody discovery and for affinity maturation of lead antibody candidates. The genetic sequence of the starting antibody is diversified into a library of variants. Each genetic variant is inserted into a phage, in turn inserted into a bacterial factory that pumps out phage replicas bearing the antibody variant in protein format.
In a high-throughput process called biopanning, the antibody target (the epitope) is immobilised and a sample of each variant in the library is washed over the target. Any variants that bind to the target are considered hits.
Phage display libraries – big but have they delivered?
The “display” principle linking genotype and phenotype isn’t limited to phage. Numerous display systems exist, where the antibody of interest is displayed on ribosomes, or on the cell surface of bacteria, yeast or mammalian cells. Each system has its pros and cons, but phage display has several advantages. First the bacterial libraries hosting the phage are relatively fast, cheap and easy to grow up, compared to slower growing yeast and mammalian cells. Phage display has another big advantage – and that is big, big libraries of up to 10^12 variants.
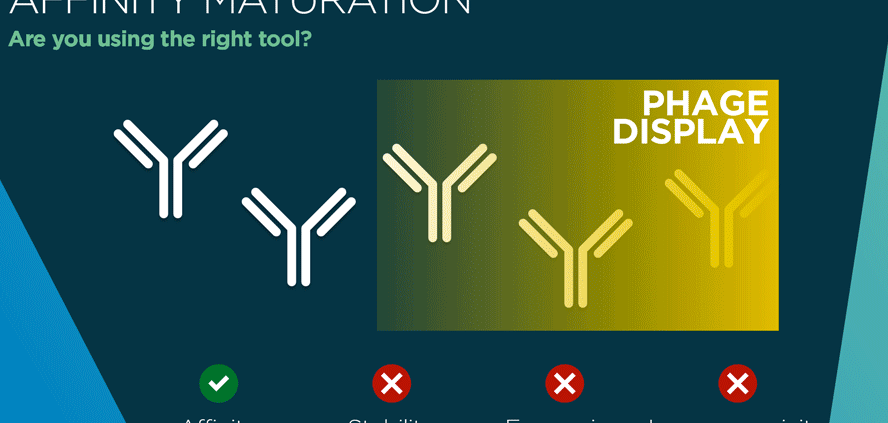
However these large phage libraries come with a price, favouring quantity over quality. While fast, synthetic and semi-synthetic methods of creating library diversity create artificial variants that would never be generated by natural B cell diversification. In addition, synthetic random libraries can be skewed with certain bases showing up too often (or not often enough) at certain positions in the DNA sequence (3). The knock-on effect is the presence of amino acids in “unnatural” spots in the antibody. Such variants may have good enough affinity to register as hits in phage display biopanning, but these “hits” may turn out to have folding or expression problems.
Express yourself
Expression bias and folding errors are two of the biggest drawbacks of phage display. The prokaryote machinery of bacteria is simply not equipped to fold complex human antibodies and cannot make crucial post-translational modifications such as forming disulphide bonds. E.coli, the most popular phage host, can be picky about which antibodies it makes, preferring to express proteins rich in certain amino acids such as methionine and lysine. Phage display handles expression and folding of smaller proteins such as functional snippets of antibodies much better. Single-chain variable fragments (scFv) of antibodies are most suited to phage display, followed by antigen-binding fragments (Fab) (4). However even scFv expression is not without its problems. For example, these antibody snippets are prone to aggregation which can cause false negatives or positives. Another limitation with antibody fragments such as scFv occurs if they need to be reformatted to full length IgG or other formats. After the high-throughput screening phase of phage display, reformatting promising scFv variants can bring an antibody development project skidding back to a crawl. Plus, a promising scFv can turn out to have disappointing affinity once the whole antibody is expressed and folded properly in a mammalian cell.
The eukaryote protein expression and post-translational machinery of yeast display can overcome some of the expression problems of prokaryote phage display. But amongst the trade-offs are smaller library sizes and the larger volumes needed for growing enough yeast cells (5).
Alternatives on display
Some of phage display’s limitations can be overcome by pairing phage display with other technologies. Some groups pair phage display with yeast display (6), or screen phage display hits for functionality with flow cytometry (7) while others batch reformat scFv to IgG before further functionality analysis. But all of these approaches take time, money and resources.
Another option is to bypass phage display altogether. Here at Fusion Antibodies, our rational affinity maturation platform (RAMPTM) sidesteps most of the issues with phage display. Our rationally designed in silico libraries are magnitudes larger than phage libraries, with around 10^25 variants. Diversity is introduced rationally, following the example of somatic hypermutation, which minimises the risk of finding amino acids in unexpected places. Rapid in silico screening of the library yields a micro-library of the strongest candidates which are then expressed directly as full length IgGs in mammalian CHO cells – neatly avoiding expression bias, folding problems and the need to reformat fragments to full IgGs. RAMP™ can also be used in combination with phage display. For example for performing affinity maturation of a phage-derived lead antibody candidate, or for sequence optimisation to improve expression and stability in CHO cells.
- https://www.ncbi.nlm.nih.gov/pmc/articles/pmid/30516432/
- https://en.wikipedia.org/wiki/Monoclonal_antibody_therapy#FDA-approved_therapeutic_antibodies
- https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0193332
- https://www.ncbi.nlm.nih.gov/pmc/articles/pmid/28613102/
- https://academic.oup.com/peds/article/24/9/711/1558039
- https://academic.oup.com/peds/article/24/9/711/1558039
- https://www.sciencedirect.com/science/article/pii/S1871678417302121
Learn how RAMPTM can boost or replace phage display for affinity maturation and sequence optimisation of your antibody